Förderzusagen
Teilnehmer PostDoc Programm
Abstract
Eosinophils are multifaceted granulocytes that play critical roles in both tissue homeostasis and disease pathology. While traditionally viewed merely as end-stage effector cells in
helminth infections and allergy, recent evidence has redefined them as versatile regulators capable of shaping adaptive immunity, tissue repair, and metabolic homeostasis. Despite this
progress, the transcriptional programs that dictate whether an eosinophil acts as a homeostatic regulator or a pathogenic effector remain poorly understood. This project focuses
on cJun, a core component of the AP-1 transcription factor complex, as a potential „master switch“ for eosinophil function. High cJun gene expression in human eosinophils correlates
with severe asthma, yet its specific role in eosinophil function has never been defined.
To isolate the function of cJun, our lab utilized Cre/LoxP technology to generate an eosinophil specific knockout mouse model (cJun EoCre). My preliminary characterization of these mice has several breakthrough observations. In steady-state conditions, immunophenotyping revealed that cJun is dispensable for eosinophil development and survival, as knockout mice show normal eosinophil numbers in the bone marrow, blood, and lung. However, these cells display a dysregulated surface phenotype, characterized by increased antigen-presentation potential and altered adhesion profiles. Furthermore, using the K/BxN serum-transfer arthritis model, I observed that cJun EoCre mice were protected from joint swelling and pathological bone destruction. Histological assessment using TRAP and H&E staining of the tibia and paw, combined with µCT analysis, revealed a significant reduction in osteoclasts and preserved bone microarchitecture in knockout mice. Importantly, this protection occurred without a reduction in eosinophil numbers, suggesting that cJun deletion specifically impairs the pathogenic competence of the eosinophil to drive tissue damage.
These findings create a compelling scientific puzzle: cJun deletion is protective in arthritis (a Type 1/17 environment), yet human data implies high cJun expression is pathogenic in asthma (a Type 2 environment). I hypothesize that cJun functions as a context-dependent transcriptional switch that is specifically required to program pathogenic eosinophil effector
functions in response to Th2 inflammatory signals. This project aims to characterize this requirement within the context of Type 2 allergic airway inflammation to identify novel
therapeutic targets for eosinophil-driven diseases.
To test this hypothesis, I will employ a comprehensive suite of established immunological and genomic methodologies. I will utilize the Ovalbumin (OVA)-induced allergic airway
inflammation model to challenge cJun EoCre and WT mice. This will involve deep phenotyping using high-parameter flow cytometry to quantify airway infiltration and assess eosinophil
polarization, distinguishing between regulatory (rEos) and inflammatory (iEos) phenotypes. Concurrently, I will quantify the expression of other Jun transcription factor members via RT
qPCR. Histological evaluation of the lungs will be performed to validate the model and detect tissue-level differences in the knockout mice, alongside ELISA quantification of cytokines
within the inflammatory environment. Additionally, I will perform transcriptional profiling via bulk RNA-sequencing on eosinophils isolated via MACS directly from the inflamed lungs to identify the gene networks controlled by cJun in situ.
These in vivo studies will be complemented by mechanistic dissection using bone marrow derived eosinophils (BMdEos) differentiated ex vivo. I will compare WT and cJun-deficient
eosinophils generated via a standardized 14-day differentiation protocol (SCF/Flt3-L expansion followed by IL-5 maturation). To quantify the impact of cJun deletion on activation,
I will challenge these cells with a prioritized panel of canonical Th2, homeostatic, and Type 1 inflammatory stimuli. Functional readout will be assessed via flow cytometry quantifying
activation markers (e.g., CD86, MHCII) and adhesion molecules, alongside a bead-based immunoassay like LEGENDplex to profile the secretion of a broad panel of inflammatory and
regulatory cytokines. To investigate cytoskeletal remodeling, I will perform high-resolution fluorescence microscopy (Leica Thunder Imager); quantitative analysis (ImageJ) of
DAPI/Phalloidin-stained cells will determine if cJun regulates the morphological plasticity required for tissue infiltration. Finally, to map the transcriptional hierarchy, I will employ a
targeted RT-qPCR screen across stimulation conditions to monitor canonical markers and the entire AP-1 family for compensatory upregulation.
Optogenetic functionalization of 5-HT receptors for receptor-specific manipulations in vivo
Abstract:
Serotonin (5-HT) is a neurotransmitter distributed across the central and peripheral nervous systems of vertebrates and invertebrates 1. 5-HT functions via 5-HT receptors, G protein-coupled receptors for which seven subtypes (5HT-1 – 5-HT-7) have been identified in vertebrates and most of which are conserved across different species including Drosophila melanogaster 2,3. 5-HT has been studied as a regulator of various behaviours including appetite, sleep regulation, memory, and learning 1,4–8. Functional and clinical studies have shown that 5-HT receptors are highly relevant for mood disorders and modulate anxiety, depression, and aggression across different species 5. However, their cell and receptor-type specific functions in regulating neuronal circuit function and animal behaviour are poorly understood. This is partly due to the complexity of serotonergic innervation, signalling, and receptor expression in mammals, and the inherent limitations in pharmacological studies to selectively manipulate 5-HT receptors with a high spatiotemporal resolution, given their diverse function and molecular characteristics.
Studies have struggled to target receptor subtype cellular-specific function using agonists and antagonists hindering further pharmacological approaches 9–11.
The recent development of optogenetic approaches offers the opportunity to fill this gap as it enables light dependent GPCR manipulation in vivo through the design of chimeric light-sensitive Rhodopsins with target receptor-specific signalling properties (optoXRs) 12. This has so far not been achieved for 5HT receptors and proper characterization of such optoXRs requires in vitro and in vivo dissection of signalling properties, protein localization, and function.
The conserved function of GPCRs across species makes Drosophila melanogaster a suitable model to characterize 5-HT receptors and corresponding optoXRs in vivo. Drosophila is a powerful model due to its known connectome (i.e. precise synaptic connectivity map) and the plethora of genetic tools available. Drosophila also offers a highly accessible system for optogenetic analysis of neuronal network activity 13. Furthermore, optogenetically functionalized Drosophila Dopamine receptors were successfully designed and validated studying cell type and
receptor-specific regulation of neuromodulatory signals in specific behavioural responses 14. This study aims at generating and characterizing novel optoXRs of conserved serotonin receptors, Drosophila 5 HT-7 and 5-HT-1B, based on an improved approach used for the opto-Dopamine receptors, taking advantage of the evolutionarily conserved interaction between Rhodopsin GPCRs and their respective heterotrimeric G proteins 12, 15. Through the further improvement and generalization of the design strategy I aim to characterise 5-HT receptors using Drosophila as a model and from there move into higher organisms. I aim to establish an easily accessible workflow of experiments to generate and characterize functional optoXRs with the incorporation of bioinformatic analyses, and in vitro, and in vivo data.
Objectives and established methods:
1. Generate a workflow of bioinformatic analysis. In collaboration with the Sticht lab (Department of Biochemistry, FAU) I am employing available bioinformatic tools for de novo structure prediction of optoXRs and structural alignment with 5-HT receptors. We are generating and optimizing the designed optoXRs similar to a sequence and structure-based approach successfully used to make optoXRs 14,15 while implementing advanced structural predictions and alignments using Alphafold3 and PyMol. Designed optoXRs are modelled with their respective
Gα-subunit against the wildtype receptor bound to the same Gα-subunit, and models are ranked depending on analytical scores showing the best alignment with the wildtype receptor.
2. Testing constructed opto5-HT receptors in vitro. OptoXRs with the highest score/best alignment are selected, synthesized and expressed in a CMV-promoter containing vector backbone to test their functionality compared to the wild-type receptor in cell culture assays. In addition to immunocytochemical approaches to confirm expression, we use two well-established assays (Gsx assay and TRUPATH) in our laboratory to confirm correct G protein coupling and signalling kinetics 14,16. The Gsx assay utilizes chimeric Gαs subunits, which can convert the interaction of opsins to diverse G proteins into increases in cAMP levels, measured with a real-time reporter in living cells. The TRUPATH assay is based on 14 optimized Bioluminescence Resonance Energy Transfer (BRET) Gαβγ biosensors. Upon activation of the receptor, the heterotrimeric subunits will dissociate and a decrease in BRET signals can be measured. The combination of Gsx and TRUPATH allows us to compare the G protein signalling properties and kinetics.
3. Construction of optoXR transgenes for conditional expression in Drosophila melanogaster. Functional opto5 HT chimera will be used for transgenesis in Drosophila as previously published 14, using site-specific transgene insertion in the Drosophila genome and allowing Gal4-dependent control of cell type-specific expression (UAS opto5-HT).
4. Expression and functional analysis. Once the UAS-opto5HT lines are produced, using the plethora of genetic tools available in Drosophila, we can further analyse and characterize their in vivo activity using fluorescent 2nd messenger reporters (cAMP, calcium) and compare expression and physiological functionality with their wild type counterparts in Drosophila. Endogenously tagged 5-HT receptor fly lines are available and in vivo imaging of cAMP and calcium responses is established 14.
5. Establish a reusable workflow. The final goal of this project is to establish a reusable and flexible workflow that can be applied to more than one GPCR class across species. We aim to establish the bioinformatic basis generating rules to optimize in silico design that can be translated to other receptors and to streamline successful optoXR generation, characterization, and in vivo use. This project is still in the beginning phases, and I am actively working on objectives 1 and 2, currently testing preliminary designs of Drosophila 5HT7 optoXRs in vitro.
Generation of active and passive vaccine candidates against Borna Disease Virus 1 (BoDV-1)
Abstract:
The Borna Disease Virus (BoDV-1) is a negative-sense, single-stranded RNA virus (order Mononegavirales, family Bornaviridae) witl1 a broad tropism for mammalian species. The viral genome encodes six proteins: the surface glycoprotein G, four structural proteins (M, N, P, X) and the viral polymerase (L). BoDV-1 has been known for causing a fatal disorder of behavior and movement in horses and sheep in endemic areas across Central Europe. The natural reservoir of BoDV-1 is the bicolored white-tootl1ed sl1rew (Crocidura Jeucodon). As of 2018, it became evident that BoDV-1 may infect humans, too, leading to fatal rneningoencephalitis in over 95 % of patients. Vaccination of cattle against BoDV-1 using a live-attenuated vaccine was cornrnon in German endernic areas in the last century. Small anirnal studies observed that the elicitation of a rapid and strong l1u111oral immune response correlates with protection frorn disease.
My project aims at generating an active BoDV-1 vaccine candidate – a recombinant nanoparticle incorporating all viral proteins and molecular adjuvants as a safe, mimetic alternative to the liveattenuated vaccine virus – as weil as a passive vaccine consisting of BoDV-1-neutralizing antibodies.
At first, stabilizing mutations used to generate native-like soluble HIV-1 Env trimers shall be applied on the BoDV-1 glycoprotein (G). Hereafter, 1 plan to express different soluble variants of G in HEK293F cells and to purify the proteins via Lectin affinity and size-exclusion chromatography. In parallel, cationic liposomes, that incorporate the BoDV-1 structural proteins as weil as rnolecular adjuvants via electrostatically-driven encapsulation, will be syntl1esized. In a final production step, purified G variants will be conjugated to tl1e nanoparticle surface via established, covalent coupling mechanisms. Tlle liposomes will tl1en be characterized by TEM, DLS and HPLC.
In an extensive in vivo immunogenicity screening, wild-type C57bl/6 mouse groups will be immunized with recombinant BoDV-1 liposomes incorporating different combinations of G variants and molecular adjuvants. After two immunizations, immune sera will be tested for antigen binding via ELISA and the top five nanoparticle variations that induced the strongest humoral immune responses will be selected for further studies.
TRIANNI mice, that harbor the entire set of human immunoglobulin V region gene segments and produce antibodies with human lg variable (V) regions and murine constant (C) regions, will be immunized with the five favorable l,iposomal BoDV-1 vaccines. As with the wildtype mice, immunogenicity will be evaluated by ELISA, especially the antibody responses against G. Additionally, tl1e sera will be tested for neutralization of a virulent BoDV-1 strain. After a tl1ird booster immunization, the B cells from TRIANNI mice with neutralizing activity will be isolated, fused with SP2/0 Ag14 hybridoma cells and seeded on 96-well plates with approximately one cell per weil. After 20 days, G-binding hybridoma clones will be selected and expanded. Following expansion, the secreted antibodies of those clones will be tested in a neutralization assay with a virulent BoDV-1 strain. The B cell receptor sequences of neutralizing B cell clones will be identified via 10x genomics single cell sequencing. The nucleotide sequences encoding the human variable region exons of the heavy and light chains will be ordered at a gene synthesis company and fused with both human and murine lgG1 heavy chain and kappa light cl1ain constant regions, respectively.
In case of promising results, 1 will prepare a TVA in collaboration with the Friedrich-Löffler-Institut aiming at the performance of preclinical challenge studies after active and passive immunization with the vaccine candidates described above.
Already established methods necessary for the project:
- Strategies to generate stabilized, recombinant forms of viral glycoprotein
- High-throughput expression and purification of viral glycoproteins
- Synthesis and functionalization of liposomes
- lncorporation of TLR ligands and promiscuous peptides into liposomes
- Access to antibody-humanized TRIANNI mice via breeding
- lrnrnunomodulatory vaccine regirnen to elicit rapid humoral immune responses Evaluation of humoral immune responses via ELISAs with immune sera
- Isolation, sorting and hybridoma fusion of primary B cells
- ldentification of B cell receptor sequences via single-cell VDJ sequencing
- Cloning and generation of fully l1uman antibody clones as passive vaccine candidates
Investigating Small Cell Lung Cancer Plasticity Using Precision-Cut Tissue Slices (PCTS)
Small cell lung cancer (SCLC) is a highly aggressive malignancy marked by rapid progression, early metastasis, and high rates of therapy resistance. Recent research has revealed that
SCLC is not a homogeneous disease but exhibits significant cellular plasticity and subtype heterogeneity, which are central to its adaptability and poor clinical outcomes. SCLC plasticity
involves transitions between neuroendocrine (NE) and non-neuroendocrine (non-NE) states, driven by factors such as NOTCH and MYC signaling, chromatin modifiers (e.g., EZH2,
KDM6A), and epigenetic dysregulation. This plasticity enables tumor cells to evade immune detection, adapt to therapy, and metastasize. Plasticity is closely linked to therapy resistance,
as SCLC cells can shift phenotypes in response to chemotherapy, leading to relapse and poor survival.
To address this gap, my research employs Precision-Cut Tissue Slices (PCTS) as a core experimental platform. PCTS preserve the native tissue architecture, stromal components,
extracellular matrix, and microenvironmental cues, allowing the study of dynamic tumorcell state changes under physiologically relevant conditions. This makes PCTS uniquely suited for
dissecting mechanisms of SCLC plasticity, including lineage transitions, microenvironment driven phenotype shifts, and treatment-induced remodeling of tumor states.
PCTS enable the study of rapid lineage transitions under controlled perturbations while maintaining physiological relevance. By integrating PCTS with imaging, immunostaining,
viability assays, and transcript-based analyses, this project seeks to define the early drivers and functional consequences of SCLC state changes. The work provides a platform for
collaboration with groups focused on tumor microenvironment, drug testing, or tumor biology of other entities that metastasize to the lung and supports the development of improved
therapeutic strategies.
Phenotypic & Functional Readouts
- Immunohistochemistry / Immunofluorescence for NE and non-NE markers, proliferation, apoptosis, and microenvironmental components.
- Viability and metabolic assays adapted for ex vivo slice cultures.
- Microscopy and imaging (fluorescence, confocal) to quantify spatial patterns, cell-state distributions, and morphological changes.
- RNAseq-based analyses of different cell populations in the PCTS cultures.
- Time-course perturbation assays to investigate kinetics of state transitions.
Model Systems
As model systems, we use immunocompetent wild type BL6 mice for PCTS cultures as well as a Cre-inducible SCLC mouse model driven by conditional deletion of RB1 and p53 extended
in vitro with murine and human SCLC cell lines. I recently established a Cre-induction method in vivo suitable for working at biological safety level S1 conditions with comparable efficacy to a viral particle Cre-delivery system published in Communications Biology (2025). This method is very valuable to study plasticity ex vivo and in vivo.
The role of Tifa in epithelial and immune cells in intestinal homeostasis and inflammation
Abstract:
Inflammatory bowel diseases (IBD), such as Crohn’s disease and ulcerative colitis, impair the lives of more than four million people worldwide. The development of effective targeted therapies is an unmet clinical need and requires a deep understanding of their underlying molecular mechanisms. Epithelial barrier dysfunction is a hallmark of intestinal inflammation and is considered a major pathogenic mechanism driving IBD.
In our study of the role of TIFA in intestinal epithelial cells (IEC), we identify TIFA as a link between the microbial metabolite ADP-heptose and epithelial innate immune responses and describe a novel and unexpected function of TIFA as a driver of intestinal inflammation in mice. We describe that TIFA is strongly induced in mice with experimental colitis and upregulated in IBD patients. We further identify IL-22 signaling via STAT3 as a key mechanism driving TIFA expression in the gut epithelium. At the molecular level, we demonstrate that TIFA expression sensitizes the gut barrier to the bacterial metabolite ADP-heptose. ADP-heptose-induced TIFA signaling orchestrates an inflammatory epithelial response with NF-κB and inflammasome activation as well as high levels of chemokine production. Finally, we provide functional evidence that mice lacking TIFA are protected from colitis development, underlining a functional role of TIFA signaling in the pathogenesis of intestinal inflammation. Collectively, these results suggest that TIFA may serve as a potential therapeutic target in gut infection and inflammation.
In our study investigating the role of TIFA in immune cells, we demonstrated that CD4+ T cells lacking TIFA induce fewer colitis symptoms in a T cell transfer colitis model. However, it remains unclear whether this reduction is due to impaired T cell migration, function or both, and requires further investigation. In addition, we found that TIFA deficiency appears to affect the polarization of T cells into Th17 cells, an observation that needs to be further investigated, as well as the effects of TIFA deficiency on other T cell subsets. With regard to B cells, our findings suggest that TIFA deficiency does not affect their maturation. However, preliminary analyses, including bulk RNA sequencing and single-cell RNA sequencing, indicate functional impairment in TIFA-deficient B cells. Interestingly, analysis of 1-year-old Tifa-/- mice revealed a pronounced immune cell infiltration exclusively in the colon, a phenotype that needs to be further investigated to identify the underlying immune cell type that triggers these effects.
Established methods:
- Mouse models:
- Tifa-/- mice
- TifaΔIEC mice (deletion of Tifa only in intestinal epithelial cells)
- TifaΔCD4 tdTomato mice (deletion of Tifa only in CD4+ T cells, which are additionally labelled with
tdTomato) - TifaΔBcells mice (deletion of Tifa only in immature and mature B cells)
- In vivo inflammation and infection models: DSS colitis, Citrobacter rodentium infection, αCD3 injection as a T cell activation model, T cell transfer colitis
- In vivo rektal administration of ADP-heptose
- In vitro cultivation of mouse and human organoids (3D and 2D monolayer cultures)
- Immunofluorescence staining and RNA Scope staining
- Tifa-mScarlet2 transfection vector
- T cell isolation and culture with in vitro T cell polarization
- Flow cytometry of T cell subsets
- Isolation of B cells
- Flow cytometry of B cell populations
Abstract
Gene regulation constitutes a fundamental determinant of biological processes, and its perturbation is a critical factor in the transition from physiological to pathological states. Elucidating gene regulatory circuits is therefore essential for understanding disease mechanisms and identifying novel therapeutic targets. At the transcriptional level, regulatory control is mediated through the coordinated interaction of cis-regulatory elements, transcription factors (TFs), and their target genes.
Computationally, these interactions are formalized as gene regulatory networks (GRNs), in which TFs, regulatory regions, and genes are represented as nodes, and their regulatory relationships are encoded as edges.
The investigation of gene regulation has long been a central objective in molecular biology, with significant insights derived from bulk-level measurements. However, the advent of single-cell technologies has transformed this field by enabling the analysis of regulatory circuits at unprecedented resolution. These approaches allow the dissection of cellular heterogeneity and the generation of high-dimensional datasets across thousands of individual cells, thereby providing a robust foundation for GRN inference through computational algorithms. In particular, the emergence of single-cell technologies such as 10x multiome sequencing, which simultaneously capture chromatin accessibility and transcriptomic profiles within the same cell, facilitates a fine-grained reconstruction of gene regulatory circuits and offers new opportunities for mechanistic discovery.
In recent years, a multitude of single-cell multiome data have been generated for healthy kidney as well as for pathological conditions, including acute kidney injury (AKI), chronic kidney disease (CKD), and renal cancer. This project aims to leverage these datasets to systematically dissect gene regulatory mechanisms within the kidney and to identify critical regulatory nodes that are perturbed in disease states. Furthermore, by integrating complementary bulk-level datasets, we will enhance the resolution and robustness of the inferred gene regulatory networks, thereby refining our understanding of kidney-specific regulatory architecture under both physiological and pathological conditions.
Established bioinformatic data analysis methods for:
- bulk RNA-seq, ChIP-seq, CUT&RUN, CUT&Tag, ATAC-seq
- scRNA-seq, scATAC-seq and single-cell multiome sequencing
- Gene Regulatory Network inference with SCENIC+
Characterization of intestinal Amyloid Precursor Protein in the pathogenesis of colorectal cancer
Abstract:
Colorectal cancer (CRC) is the second most commonly diagnosed cancer and the second most common cause of cancer-related death in Europe. More than 57,000 new cases of CRC are diagnosed each year in Germany. Although early detection methods have reduced incidence and mortality, treatment remains a challenge due to the problems associated with chemotherapy resistance and the high mortality rate of metastatic CRC. Therefore, there is an urgent clinical need to develop new therapeutic approaches to combat CRC.
Amyloid precursor protein (APP) was identified as a component of the neuritic plaques that are a hallmark of Alzheimer’s disease (AD). Further research on APP, particularly in the neurosciences, has shed light on its expression and dynamics. In neurons, APP has been described as a transmembrane protein that undergoes two sequential cleavages to produce different peptides. In the first step, APP is cleaved by either alpha- or beta-secretases. When an alpha (α) secretase catalyses the first step, it produces two fragments, soluble APPα (sAPPα) and the membrane-bound peptide C83. In contrast, when this first step is catalysed by the beta (β) secretase BACE1, the processing of APP produces sAPPβ and C99. The second cleavage is then carried out by gamma (γ) secretase, with presenilin 1 as the catalytic centre. From C83, the γ-secretase complex releases P3 and AICD (APP intracellular domain), whereas from C99 it produces Aβ (the peptide that forms senile plaques) and AICD. In a very recent study, we were able to show that, in addition to the brain, Presenilin 1 is highly expressed in the intestinal epithelium. Surprisingly, we were also able to show that Presenilin 1 is not required for normal intestinal development, but was identified as a key driver of intestinal tumor development. Given this unexpected finding, there is a strong rationale for investigating the involvement of APP and its cleavage products in intestinal tumors. We observed that APP is highly expressed in the intestinal epithelium in both mouse and human intestinal tissues. Furthermore, the expression of APP and its processing are increased during intestinal tumorigenesis. Finally, preliminary functional data show that deletion of APP in colon cancer cell lines sensitises them to apoptosis in vitro.
Based on our preliminary data, we hypothesise that APP has an impact on intestinal tumor development and that modulation of APP or its processing may have therapeutic potential in colorectal cancer. To test this hypothesis, we will determine the role of epithelial APP in intestinal tumorigenesis in vivo and decipher APP-regulated pathways in intestinal epithelial tumor cells. We will also characterise APP expression and processing in human samples. Finally, we will investigate the therapeutic potential of APP in this disease.
Established methods:
- Mouse models (AppΔIEC , App/Aplp2ΔIEC)
- Experimental induction of colorectal cancer (AOM/DSS, Apcmin/+, tumor xenotransplantation)
- In vitro cultivation of tumor organoids derived from human and mouse tumor cells (2D and 3D)
- RNAscope and immunofluorescence stainings
- Generation of knockout cell lines and organoids using CRISPR-Cas9 technology
- Flow cytometry
Extracorporeal photopheresis induces a tolerogenic phenotype in primary human blood DCs
Abstract:
Graft-versus-host-disease (GvHD) and allograft rejection are complications observed in patients undergoing hematopoietic stem cell therapy or solid organ transplantation, respectively.
Immunosuppressive regimens are the first-line therapy but not always effective and inducing strong side effects in patients suffering from GvHD and allograft rejection. Extracorporeal photopheresis (ECP) represents a second-line therapy for these complications. For ECP-treatment, blood leukocytes are collected by apheresis and treated ex vivo with the photosensitizer 8-Methoxypsoralen (8-MOP) followed by irradiation with ultraviolet A light (UV-A). Subsequently, cells are transfused back into the patient. ECP shows promising treatment results especially in chronic GvHD with minimal side effects. However, the mode of action of ECP is still not completely understood. It is known that the majority of T cells undergo apoptosis during treatment with ECP, whereas monocytes are resistant to cell death but differentiate into dendritic cells (DCs). Further, murine GvHD models showed that transfer of ECP treated CD11c+ DCs is sufficient to mediate the effect of ECP. Thus, DCs seem to have a crucial role in the induction of immune tolerance during ECP treatment. However, it is unknown how primary human DC subpopulations respond to ECP, i.e. whether ECP induces cell death or an immunomodulatory phenotype in DCs. In humans, DCs are defined by lack of expression of so-called lineage markers (CD3 for T cells, CD19/CD20 for B cells, CD14 for monocytes, and CD56/CD335 for NK cells) but high expression of HLA-DR, the marker for professional antigen-presenting cells. Based on ontogeny and surface markers, DCs can be distinguished into conventional DC type 1 (cDC1), DC2, DC3, and plasmacytoid DCs (pDC). While cDC1, DC2, and DC3 are known for their capacity to induce strong T cell responses, pDC can produce vast amounts of type I interferon in response to viruses. As DCs are the most efficient inducers of T cell immune responses, they are very important for anti-viral and anti tumor immunity. However, DCs have also a central role in immune tolerance, as they can induce and expand regulatory T cells and lead to anergy in autoreactive T cells.
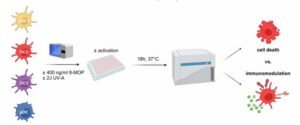
Due to their important role in the regulation of immune responses, we hypothesize that ECP induces a tolerogenic phenotype in human DCs, which contributes to the mode of action of ECP in GvHD patients. Therefore, we are analyzing how the primary human DC subpopulations cDC1, DC2, DC3, and pDC react to in vitro treatment with ECP. As DCs are very rare (representing less than 1% of peripheral blood mononuclear cells (PBMCs)), the different DC subpopulations are isolated by cell-sorting from PBMCs of healthy blood donors after negative enrichment using magnetic beads. Subsequently, cells are incubated with 8-MOP and irradiated with UV-A light in steady state as well as under inflammatory conditions and analyzed for viability and their phenotype by different flow cytometry-based assays (see Figure 1). As T cells undergo cell death after ECP-treatment, we analyze cell death by staining for Annexin V as well as 7-AAD to differentiate between apoptosis and necrosis. Further, we analyze the expression of co-stimulatory as well as co-regulatory molecules by flow cytometry. Additionally, we collect the supernatants of the treated DCs to measure secreted inflammatory and tolerogenic cytokines.
Thereby, we are able to determine whether ECP induces cell death or an immunomodulatory phenotype in DCs. The experiments are also performed with DCs stimulated with Toll-like receptor ligands to mirror the inflammatory environment in GvHD patients. In a next step, ECP-treated DCs will be co-cultured with allogeneic naïve (mixed leukocyte reaction) or autologous memory (antigen-specific T cell assays) T cells. These experiments will show whether ECP-treated DCs are able to induce T cell response and whether ECP polarizes the T cell response towards regulatory T cells.
Figure 1. Schematic overview for in vitro treatment of sorted human blood DCs with ECP.
As during ECP only a minority of the blood immune cells of the patient is directly treated, ECP might also have indirect effects on DCs. As ECP induces apoptosis in T cells as well as other immune cells and DCs are able to recognize and phagocytose apoptotic cells via specific surface receptors such as TIM-3, we hypothesize that ECP-treatment induces a tolerogenic phenotype in DCs via an indirect pathway. Therefore, purified DC subpopulations will be cultured with ECP-treated PBMCs in order to analyze the effect of apoptotic cells, which arise under ECP-treatment, on the phenotype and function of DCs. In order to determine whether DCs phagocytose ECP-treated cells, PBMCs will be labelled with CFSE prior to ECP-treatment. As described above, we will determine the viability and phenotype of the DCs as well as determine cytokine secretion by CBA assays. Thereby, we will determine whether human primary DCs directly or indirectly respond to treatment with ECP and might be involved in the therapeutic effect observed in patients.
Established methods:
- Isolation of immune cells from different human tissues
- Blood, spleen, thymus, lymph nodes, tonsils, bone marrow, breast and breast cancer tissue, lung
- High-dimensional flow cytometry (BD LSRFortessa, Beckman Coulter CytoFlex S)
- Flow cytometry-based cell sorting (BD Aria II cell sorter SORP)
- Six-color confocal immunofluorescence microscopy (Zeiss LSM780)
- Cytometric Bead Array (BD FlexSets, BioLegend LEGENDplex)
- Cell death assays (LDH cytotoxicity assays, 7-AAD vs. Annexin V)
- DC differentiation assays on MS-5 stroma cells
- DC:T cell co-cultures (mixed leukocyte reactions, antigen-specific memory assays)
- Inflammasome activation (Caspase 1 activity, inhibitors)
Interplay of epithelial mechanics and neutrophil activation as a pathomechanism in refractory IBD and colorectal cancer patients
Abstract:
Inflammatory bowel diseases (IBD) are a heterogeneous group of disorders characterized by gastrointestinal tract inflammation, being Crohn’s Disease and Ulcerative Colitis, the most clinically abundant. IBD affects around 6.8 million people all over the world, being Europe the continent with the highest prevalence. Although anti-TNF-α monoclonal antibodies are a breakthrough therapy, almost 40% of IBD patients do not respond to therapy or acquire resistance. This leads to uncontrolled inflammation that, prolonged in time, constitutes a higher colorectal cancer risk. This justifies the current efforts to identify innovative therapy targets for IBD understanding of the molecular processes behind (personalized medicine). The fact that an increased intestinal permeability precedes the onset of inflammation possesses epithelial disruption as a potential etiological factor in IBD. Moreover, in recent years, several publications have highlighted that an impaired epithelial barrier is linked to resistance to therapy. Nevertheless, how altered epithelial mechanics can influence the structural properties of the extracellular matrix and immune cells migrating through it is not fully understood. On the other hand, the role of neutrophils as resistance-to-therapy players has emerged very recently. Then, we hypothesize that an alteration in epithelial cell mechanics constitutes a primary event that impacts the proliferation and behaviour of the first immune cell responders: neutrophils. This would
further contribute to ECM remodelling and fibroblasts polarization to Inflammatory associated fibroblasts (another mechanism implicated in therapy resistance). Then, this project aims to elucidate how an altered epithelial dynamic constitutes a primary event that directly (through an altered transepithelial migration) or indirectly (affecting the mechanical framework) could promote the accumulation of activated neutrophils.
Established methods:
- Preclinical models of IBD (mouse lines)
- Isolation and culture of murine organoids (2D and 3D)
- Isolation and culture of human organoids (3D culture)
- Immunofluorescence and confocal laser scanning microscopy
- Isolation and culture of neutrophils from the bone-marrow and the peritoneum (mouse model of peritonitis)
- Flow cytometry of immune and stromal cell populations
- 3D-Traction force microscopy (TFM) – In collaboration with Prof. Ben Fabry and MSc. David Böhringer
- Label-free Multiphoton Imaging – In collaboration with Prof. Maximilian Waldner, Prof. Sebastian Schürmann, Dr. rer nat. Oana Maria Thoma
- Brillouin microscopy – In collaboration with Dr. Jochen Gück and Dr. Alice Battistella
Abstract:
Parkinson’s Disease (PD) is characterized by the loss of dopaminergic neurons in the substantia nigra (SN), which project to the striatum. PD and other synucleinopathies are associated with elevated levels and/or mutations in the presynaptic protein alpha-synuclein (aSyn). The misfolding and intracellular accumulation of alpha-synuclein into structures known as Lewy Bodies (LB) is a key feature in the pathogenesis of synucleinopathies, including Parkinson’s disease (PD). Phosphorylation at serine 129 of alpha Synuclein (pSer129-aSyn) is a hallmark of LB and is significantly enriched in these pathological structures, whereas only 4% of the wild type alpha-synuclein pool is phosphorylated. Despite extensive investigation, the molecular mechanisms by which quantitative or qualitative changes in aSyn lead to neurodegeneration remain poorly understood. In this project we hypothesize that phosphorylation and early increases in aSyn levels promote pathological protein-protein interactions (PPIs), which may drive disease progression. Identifying and targeting these PPIs could offer novel therapeutic strategies for PD and related
synucleinopathies. To explore this, we developed an in vivo proximity biotinylation assay to map the aSyn and pSer129-Syn interactome in HEK293T cells and in LUHMES cells that can be differentiated to post-mitotic cells with biochemical, morphological and functional features of dopaminergic (DAergic) neurons. By fusing aSyn to a promiscuous ascorbate peroxidase (APEX2) and supplementing with biotin-phenol and H2O2, proximal stable and transient interactors are biotinylated, purified via affinity methods, and identified through liquid chromatography-mass spectrometry (LC-MS). The proteins identified as interactors will further be investigated in iPSC derived dopaminergic neurons from both isogenic and SNCA gene-triplication.
Established methods:
- Plasmid cloning
- Different versions of SNCA and APEX2 (control, wildtype, phosphorylated and phospho resistant) was cloned into the expression vector pCAG-P2A-Puromicin for lentiviral
transduction into Hek293T, LUHMES and human iPSCs. - Cell line generation
Hek293T cells stably expressing control or wild type or phosphorylated or phospho-resistant aSyn tagged to APEX2 is generated. LUHMES cells expressing various version of aSyn are in
progress. The differentiation protocol of LUHMES cells to dopaminergic neuron-like cells is established. - APEX2-based proximity labeling
APEX2-based proximity labeling protocol of aSyn expressing Hek293T cells in vivo in the presence of biotin-phenol and H2O2 is optimized. The proteins labeled are enriched using
Streptavidin beads and subjected to Western blotting.
Composition and function of mGuR7 dimers at the inner hair cell ribbon synapses in the cochlea
Abstract:
In the auditory pathway, sound encoding is ensured via synaptic transmission by specialized chemical synapses, so called ribbon synapses at inner hair cells (IHC) that contact post synaptic spiral ganglion neurons. Toxic environmental stimuli such as noise trauma can lead to excessive glutamate concentrations in the synaptic cleft. This in turn might result in degeneration and loss of these sensory synapses that ultimately leads to varying forms of hearing impairment. However, G protein-coupled receptors such as metabotropic glutamate receptors (mGluR) can couple to inhibitory signal pathways that limit pre-synaptic glutamate release and thereby protect the synapse from toxic stimuli. To date, eight known mGluR types have been identified that are classified into three groups based on sequence homology, second messenger coupling and pharmacological characteristics: group I receptors (mGluR1 and 5) are typically expressed post-synaptically, whereas group II receptors (mGluR2 and 3) have been reported at pre- and post-synaptic sites. Group III receptors (mGluR4, 6, 7 and 8) show a clear preference for the pre-synaptic terminal, except for mGluR6. Especially members of group II and III have been associated with protective functions. Furthermore, several mGluR members can be alternatively spliced at the intracellular C-terminus to generate isoforms. In general, mGluRs can assemble into homo- or heterodimers.
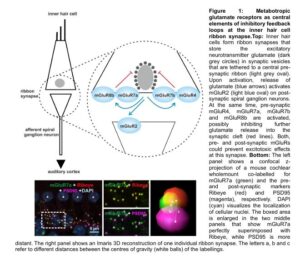
In preliminary work, I described the synaptic localization of group II and III mGluRs at inner hair cell ribbon synapses in the cochlea in detail and started to investigate possible dimer formations between detected mGluR types. By using anatomical methods such as confocal and STED microscopy, a pre-synaptic localization of mGluR4, two mGluR7 isoforms (mGluR7a
and mGluR7b) and mGluR8b was revealed, whereas mGluR2 was detected post-synaptically (Fig.1)
Interestingly, mGluR7 is the only mGluR type that has been correlated with age-related and noise‐induced hearing deficits for over ten years. Furthermore, we observed a reduction of both mGluR7 isoforms in aged animals as well as at higher frequencies in the cochlea. Most mGluR types are activated in the low micromolar range, however mGluR7 isoforms are intriguing outliers as their glutamate affinity are ~4000 fold lower compared to other mGluRs. As mGluR7 is activated only at high glutamate concentrations, the receptor could function as an intrinsic synaptic break for excessive glutamate release. In 2012 the group of Jean- Phillipe Pin demonstrated the formation of between group I mGluRs and within members of group II and III in cell culture. Moreover, overlapping expression patterns of group II and III mGluRs have been observed in the brain and indeed, mGluR2/3, mGluR2/4 and mGluR2/7 heterodimers have been recently reported in cultured neurons and in several brain regions e.g. in the cortex and the hippocampus. Recent studies described that heterodimeric mGluRs have distinct characteristics in their kinetics, glutamate affinity, receptor targeting and coupling efficacy to signal pathways compared to their homodimeric counterpart. The recently demonstrated mGluR2/7 heterodimer can be activated by group II and III agonists and shows an even higher glutamate affinity than the homodimeric mGluR2.
To understand the function of mGluR7 receptors at the IHC ribbon synapses it is necessary to analyse if they are present as homodimeric receptors or heterodimers with mGluR4 or mGluR8b. Therefore, we are – using a set of complementary anatomical, biochemical and pharmacological tools. In addition, we plan to study their function by a combination of electrophysiological and physiological techniques. During my ELAN pilot project funding two methods were established to analyse the properties of mGluR7 heterodimers: time-resolved FRET (TR-FRET) and electrophysiology. In cooperation with the group of Jean-Phillippe Pin (Montpellier, France), time-resolved FRET measurements were performed that analyse drug response curves of heterodimeric mGluRs. This technique can be complemented with immunohistochemical costainings and by co-immunoprecipitations. Obtained data will be used to test the influence of mGluR agonists on calcium currents by whole-patch clamp recordings of inner hair cells in cochlear wholemount preparations (I learned this technique during a 5 week research stay in Göttingen in the laboratory of Tobias Moser).
So far, cochlear alterations and/or hearing deficits of mGluR7 knockout animals were not analysed. Given the association of hearing deficits of mGluR7 in humans and my preliminary data e.g. presynaptic localization of mGluR7 isoforms and a reduced expression in aged animals as well as in higher encoding frequencies this is an intriguing question. Interestingly, a recent study hypothesized that differences in the behaviour of mGluR7 knockout mice might be simply caused by hearing deficits. Therefore, we plan to compare the hearing phenotype of mGluR7 knockout animals with wildtype littermates. Auditory brainstem responses (ABR) and acoustic startle responses (ASR) will be recorded in collaboration with the laboratory of Holger Schulze (Experimental Otolaryngology, FAU) who has a strong and well documented experience in a complete set of physiological and behavioural tests to analyse the auditory system in rodents.
Crosstalk between T cells subtypes and fibroblast subsets in skin autoimmunity
Autoimmune skin diseases are characterised by an aberrant immune response in the skin tissue. This involves a complex interplay of immune cells, keratinocytes, and fibroblasts, in response to specific antigens [1]. Despite the identification of several potential mediators of skin autoimmunity (altered microbiome, inherited dysfunctional immunity, antigens activating innate immunity, epigenetic modifications, sex predisposition, neoantigens and molecular mimicry), the precise mechanisms underlying skin autoimmunity remain unclear [1]. Systemic sclerosis (SSc) and systemic lupus erythematosus (SLE) are two significant skin autoimmune disorders. Although they possess distinctive characteristics and pathophysiology, they both demonstrate pronounced cutaneous involvement.
SSc is a rare, chronic, immune-mediated disease characterised by skin fibrosis. Some patients with SSc develop life-threatening systemic manifestations. The most prominent feature of SSc is the progressive fibrosis that results from an excessive accumulation of extracellular matrix (ECM) [2]. The pathogenesis of SSc is a complex and incompletely understood process involving activation of the immune system, development of vasculopathy and fibroblast activation leading to fibrosis [3]. As fibroblasts are the primary source of ECM, they are also recognised as the principal mediators of the pathogenic fibrotic tissue remodelling, also in SSc [3]. However, they also perform fundamental functions in the regulation of the immune response in autoimmune diseases, particularly interacting with T lymphocytes, not only in SSc but also in SLE [4].
SLE is an autoimmune disease with a heterogeneous clinical presentation, ranging from cutaneous to systemic organ involvement [5], affecting more than 3.4 million individuals worldwide [6]. Despite novel therapies, mortality rates remain high. Alterations in T cell differentiation and subsets have been observed in SLE patients [5]. It has been demonstrated that fibroblasts display disparate responses to inflammatory cytokines contingent depending on the specific type of lupus skin lesion [7]. Nevertheless, the mechanisms and relationships between fibroblasts and T cells in SLE remain poorly understood.
Despite existing knowledge regarding the interactions between T cells and fibroblasts, no studies have been conducted to investigate the spatial association between the different fibroblast subtypes and T cell subsets in skin autoimmunity. Furthermore, no studies have compared autoimmune diseases with and without a primary fibrotic component, as exemplified by SSc and SLE, respectively.
In a recent study, we used Imaging Mass Cytometry (IMC) to analyse skin biopsies from healthy controls and SSc patients in order to decipher the skin fibroblast heterogeneity [8]. IMC is a technique that allows the use of up to 40 metal-conjugated antibodies in a skin formalin-fixed-paraffin-embedded (FFPE) histology sample, facilitating the generation of a high-multiplex image. Consequently, it is possible to perform a comprehensive phenotyping and examine the localisations and spatial relationships between the different cell types [9]. We have identified 13 different skin fibroblast subsets, of which five were increased in SSc and three were decreased. We then investigated the spatial localization of these subsets: TFAMhigh and ADAM12+GLI1+ fibroblasts are mainly located in the upper dermis; other subsets, such as myofibroblasts or FAPhigh, are exclusively found in the lower dermis; finally, S1PR+ and THY1+ADAM12highPU.1high are the only two fibroblast subsets that are increased in both dermal layers in SSc. Lastly, we show that he percentage of S1PR+ fibroblasts as well as their percentage neighbouring the ADAM12+GLI1+ fibroblasts are significantly associated with clinical outcomes of SSc (modified Rodnan skin score). This illustrates that not only are the cell subsets of significance in the development of the disease, but also their spatial relationships.
However, pathophysiological relevance of the interactions of these fibroblast subpopulations and the T cell subsets remains unknown. The aim of the current project is to elucidate the spatial relationships between the various fibroblast subsets and T cells in skin autoimmunity and compare autoimmune diseases with and without a primary fibrotic component, as SSc and SLE respectively. As the spatial localization and relationships are basic to understand these interactions, the integration of spatial proteomics (IMC) and spatial transciptomics (Merscope) is the most suitable approach. We are establishing a 40 metal-conjugated antibody IMC panel targeting fibroblast protein markers to identify the aforementioned fibroblast subsets as well as different T cell subsets (Th1, Th2, Th17, Treg, memory and effector T cells). In a second phase of the study, spatial transcriptomic (Merscope technology) will be used to ascertain the specific molecular mechanisms involved in these cell-to-cell interactions. A Merscope panel with 500 gene will be designed. It will include all the genes encoding the protein markers used in the IMC panel and markers to deeper phenotype the fibroblasts and T cells. Additionally, we will also add gene probes to elucidate the molecules involved: cytokines, chemokines and chemokines receptors and ligand-receptor partners, among others.
In collaboration with Dr. med. Christina Bergmann (University Clinic Medicine 3), our group has been collecting skin FFPE histology samples from patients diagnosed with SSc and age- and sex-matched healthy donors. In collaboration with Dr. med. Panagiotis Garantziotis (University Clinic Medicine 3) we are starting to collect SLE skin biopsies. When possible, biopsies from affected and non-affected skin are collected.
In the last years, the applicant has acquired expertise in IMC panel design, established SOPs for IMC experiments and developed the bioinformatics analysis pipelines based on the approach originally proposed by the Bodenmiller group at the University of Zurich [10]. These pipelines are intended for the analysis of multiplex, high-dimensional images. Briefly: using the Python based software Steinbock, the raw IMC data are converted to TIFF files, followed by the cell segmentation to obtain the single cell data; cells of interest are selected using the flow cytometry analysis software FlowJo®; applying the R package imcRtools is used to perform the first staining quality control, as well as dimensionality reduction, Phenograph and FlowSOM clustering and the spatial analysis (interaction and neighbourhood). We plan to use these methods to elucidate the cellular relationships between the fibroblast subpopulations and T cell subsets.
The Department of Internal Medicine 3 has available the spatial transcriptomics Merscope platform (MERFISH). The applicant has optimised SOPs for the utilisation of FFPE samples. This technique enables the study of the single-cell transcriptome in a tissue sample without the need for tissue digestion, thus preserving the tissue structure and the cell localisation. An approach similar to the IMC analysis will be applied to the new data in order to unravel the cell-to-cell molecular mechanisms.
The combination of these two techniques allows us to uncover the cellular crosstalk between fibroblast subsets and T cell subtypes. This is accomplished by identifying two essential aspects: firstly, which cells are in close proximity to each other by IMC, and secondly, the specific molecular mediators involved in this relationship using Merscope.
Abstract:
Characteriza on of the role of microRNAs in spa otemporal pa erning of the vertebrate spinal cord In the developing spinal cord, spa ally defined classes of neurons are par oned into molecularly and func onally dis nct subtypes by a shared temporal pa erning program. This program relies on a sequen al expression of temporal transcrip on factors (tTFs) in neurons generated at successive developmental stages: early-born neurons express Onecut-family TFs, intermediate neurons express Pou2f2 and Z x2-4, and late-born neurons express Nfia/b/x and Neurod2/6. While the sequence of tTFs expression is well described, the molecular mechanisms orchestra ng this chronologically ordered program remain largely unknown. Besides possible contribu ons from cell-extrinsic signaling pathways, cell-intrinsic gene regulatory mechanisms and epigene c programs are thought to play cri cal roles in this process. Recent work in the vertebrate re na and cortex has highlighted microRNAs (miRNAs) as important regulators of temporal pa erning. Building on this, we inves gated whether miRNAs also contribute to temporal pa erning in the vertebrate spinal cord. By condi onally dele ng Dgcr8, a core component of the microprocessor complex essen al for miRNA biogenesis, in the central nervous system, we observed a complete loss of late temporal iden ty neurons accompanied by a concomitant increase in early and intermediate iden ty neurons. This phenotype underscores the crucial role of miRNAs in ensuring the correct progression of temporal iden es during spinal cord development. To test this hypothesis, I first aim to comprehensively characterize the transcrip onal changes in neuron and progenitor cell popula ons throughout the spinal cord in the Dgcr8 mutants by single cell RNA sequencing. Candidate microRNAs that may underlie such transcrip onal changes, including miR-9 and let-7, which I have already iden fied by computa onal analysis as poten al candidates involved in temporal pa erning, will then be further analyzed using in ovo electropora on and a stem cell-based in vitro differen a on model. Both miRNA families promote the specifica on of late neural iden es by antagonizing transcrip onal programs ac ve in early progenitors and neurons. Suppor ng this hypothesis, expression levels of let-7 and miR-9 increase during the neurogenic phase in the embryonic mouse spinal cord and in stem cell-based in vitro differen a on systems that recapitulate in vivo temporal pa erning. Furthermore, preliminary results of ectopic expression of these miRNAs in chicken embryos induced a premature switch from early to late neuronal iden es, providing func onal evidence for their involvement in temporal iden ty specifica on. Furthermore, preliminary data further suggest that miR-9 may regulate temporal pa erning in a dose dependent manner, a hypothesis that will be tested through targeted dele on of individual miR-9 host genes. To alter the sensi vity of tTF transcripts to miR-9, I will delete parts of the 3’-UTRs of miR-9 target tTFs, to progressively reduce the number of miR-9 sites in these transcripts. Together, this
project aims to assess the poten al role for miRNAs in orchestra ng the temporal pa erning program of the developing spinal cord. A detailed understanding of this regulatory mechanism not only advances our knowledge of spinal cord development but may also inform the ra onal design of differen a on and reprogramming protocols to generate specific neuronal subtypes for disease modeling and regenera ve medicine applica ons. Already established methods:
Dgcr8 mutant mouse model, and single-cell RNA sequencing analyses are rou nely performed in the lab. Addi onally, we have access to the stem cell-based in vitro differen a on system that effec vely recapitulates the temporal pa erning observed in vivo. I also have extensive experience with the chicken in ovo electropora on model and successfully re-established this technique in the lab upon my arrival. Furthermore, we are equipped with a comprehensive collec on of validated an bodies and established RNAscope assays, which will be used for the various phenotypic analyses required in this study.
Prospective feasibility study for deep learning-based MR-only brain radiotherapy
Radiotherapy is crucial in the treatment of malignant primary brain tumors and brain metastases. Computed tomography (CT) forms the basis for the physical dose calculation. The Hounsfield units on which the CT is based can be converted into an electron density distribution in order to model the interaction of the high-energy therapy radiation with the human tissue and thus calculate the dose to be applied to the individual patient. However, due to the very low soft tissue contrast of the CT in the brain, it is not possible to adequately define the volume to be treated or the neighboring organs at risk (brain stem, optical nerves, chiasm, etc.). An additional magnetic resonance imaging (MRI) scan is required to contour the different volumes. However, in order to provide this information during treatment planning, the contoured organs at risk and target volumes must be transferred to the CT.
The necessary rigid registration between the two image modalities is prone to errors and can lead to a systematic error of up to 2 mm [1]. This has a crucial influence on all subsequent steps in the radiotherapy treatment chain. Especially in the stereotactic treatment of brain metastases, high precision is required for the application of high doses in one (radiosurgery) or several (fractionated stereotactic treatment) treatments. In the worst case, the smallest errors in the target volume due to the transfer between CT and MRI can lead to the metastasis being missed during treatment on the linear accelerator and the entire dose being applied to healthy tissue. To compensate for these uncertainties, safety margins of a few millimeters are added to the contoured tumor or contoured metastasis as standard. However, these safety margins are limited due to the surrounding healthy brain tissue.
One way of eliminating these registration-based errors is to use an MR-only workflow. Using an additional MR sequence (acquisition time below 4min), a synthetic CT (sCT) can be generated based on artificial intelligence (AI), which contains the electron density and can therefore serve as the basis for the dose calculation. Another advantage is that only a single image modality, namely MRI, is required. This would significantly reduce time and effort, and most important the risk of registration errors. By eliminating the additional CT, which is only required for dose calculation and not for diagnostics, unnecessary additional radiation exposure can be avoided. It can be assumed that an increase in the precision and quality of treatment could lead to an improvement in the patient’s quality of life and prognosis.
Previous retrospective studies have shown that AI-based algorithms to generate synthetic CTs matches well with the dose calculation on conventional planning CTs. The deviations in the target volume were <1% [2, 3, 4]. A crucial point for the precise application of the dose on the linear accelerator is the exact positioning of the patient as during the acquisition of the planning CT, which was used as the basis for the dose calculation. Verification and correction of the patient position is carried out for primary brain tumors or brain metastases using rigid registration of 2D/2D X-ray images of the planning CT and the X-ray images taken on the accelerator before treatment. Here, too, there was good agreement when the synthetically generated CT was used instead of the standard planning CT. The maximum deviations were 3 mm for the translations and 3% for the rotations.
Although various retrospective studies show good results with the use of synthetically generated CTs, these MR-only workflows are still hardly widespread in routine practice. One aspect of this could be the limited availability or limited access to MRI scanners for radiotherapy. Furthermore, implementation in everyday clinical practice is very complex, time-consuming and requires expert knowledge in the field of MR imaging.
In a prospective study, we currently investigate the feasibility of this MR-only workflow in everyday clinical practice for all steps of the radiotherapy chain in the treatment of brain lesions. For this purpose, it must be verified whether MR imaging is possible with the necessary positioning aids used in radiotherapy. Furthermore, it must be ensured that there is dosimetric equivalence between the synthetic CT and the standard planning CT so that safe and correct dose application can be guaranteed. The virtual X-ray images generated from the sCT for positioning the patient on the linear accelerator and patient-specific quality assurance must also be verified. The study aims to validate these points and to help determine possible difficulties and exclusion criteria for patients or treatment indications. The study plan was approved by the Ethics Committee of the Medical Faculty of Friedrich-Alexander Universität Erlangen-Nürnberg (ID: 23-286-Bm) and has been registered prospectively at Clinical trials.gov (NTC06106997). Patient enrollment is expected to be completed in summer 2025. Subsequent data analysis, which will address several secondary endpoints, will require extensive work, e.g. on the patient’s movement in the thermoplastic mask during MRI acquisition.
At the same time, we are working on the development of phantoms that are suitable for regular quality assurance of this AI-based algorithm and fulfil the requirements of radiotherapy to ensure precise and safe treatment of patients. Such phantoms are currently not commercially available. The retrospective studies as well as the preliminary analyses of the prospective study show promising results for dose calculation and patient positioning at the linear accelerator with the synthetically generated CTs. In the future, the use of sCT is to be established and quality-assured for other body regions, e.g. the pelvis, in order to ensure that a larger number of patients benefit from the avoidance of registration errors and unnecessary dose exposure.
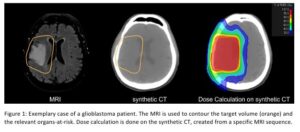
Figure 1: Exemplary case of a glioblastoma patient. The MRI is used to contour the target volume (orange) and the relevant organs-at-risk. Dose calculation is done on the synthetic CT, created from a specific MRI sequence.
Contact: Juliane SzkitsakExploring Manduca sexta larvae as an alternative model for translational imaging research
Introduction:
The Manduca sexta larva represents a suitable alternative animal model for the in vivo study of the digestive tract, due to the relatively large size of the caterpillar’s gut, with a volume of up to 1.5 ml1, comparable to that of the mouse gut. In addition, conserved disease genes, with up to 92% homology between mammals and insects2 allow the investigation of intestinal diseases in M. sexta larvae and the evaluation of imaging modalities. The primary objective of this project is to identify a biomarker for inflammatory bowel disease (IBD). Fluorescent labeling of the marker would allow the localization and monitoring of IBD in a non-invasive manner (Fig. 1A). Windfelder et al.3 identified DUOX-activation as a potential inducer of gut inflammation in
M. sexta, which is in accordance with the known overexpression of DUOX in IBD and the loss-of-function of DUOX in very-early-onset (VEO)-IBD4. This makes M. sexta a promising, alternative animal model.
Established methods:
Initial experiments to determine the suitability of various techniques for use with M. sexta larvae have already been performed. First, we established a larva DSS model to induce intestinal inflammation. Next, we established larva endoscopy with a small-animal endoscope to visualize the insect gut and to investigate inflammatory processes. To localize proteins and cells of interest in parallel with the gathering of entire structure information by imaging the unaltered structure free of artefacts, light sheet microscopy (with fixed animals) and multi-spectral-optoacoustic-tomography (MSOT) (non-invasive in live animals) were performed. Cell culture studies and immunohistochemical analysis are planned to correlate the overexpression of inflammation-induced proteins in M. sexta and humans.
Investigating the role of CXCR4 in neutrophils in development of arthritis and neutrophils reverse migration
Background:
Rheumatoid arthritis (RA) is an autoimmune disease characterized by chronic synovitis and bone erosion. Neutrophils are believed to contribute to RA as inflammatory effector cells. Hence, selective neutrophil targeting might be able to reduce arthritis. Our previous results demonstrated that neutrophils in joint of RA patients display increased transcript and protein levels of the C-X-C chemokine receptor 4 (CXCR4).
Methods:
We will study the role of CXCR4 in neutrophils in the pathogenesis of inflammatory arthritis. We will test how CXCR4 antagonists and conditional knock-out in neutrophils affect the severity of inflammatory arthritis in mice. Subsequently, we will perform detailed phenotyping and functional analysis of neutrophil populations to understand how CXCR4+ neutrophils are involved in arthritis. As CXCR4 is also highly expressed in neutrophils in the bone marrow, we hypothesize that high levels of CXCR4 on joint neutrophils are linked to reverse migration of neutrophils back and homing to bone marrow. To track the dynamic migration of neutrophils, we will leverage the transgenic ‘Kaede’ mouse strain, expressing the Kaede protein which can be converted from green (FITC) to red (PE) via violet light. Thereby, we will initially label cells in joints by violet LED lamp and track PE+ cells in other tissues including bone marrow to test the hypothesis of reverse migration of neutrophils from joint. To avoid mislabeling of cells in circulation, additional markers characterizing joint resident neutrophils (MHC II, CD64, ICAM-1) will be employed. In addition, the association between CXCR4 and reverse migration will also be examined.
Contact: Liang Zhang