Research interest
The Dulin lab is fascinated by cellular and viral processes involved in RNA synthesis. To investigate RNA synthesis, we use single molecule techniques, such as magnetic tweezers and single molecule fluorescence resonance energy transfer (FRET), as described in Single molecule biophysics and techniques.
Cellular transcription
In the Central dogma of molecular biology, RNA production is the intermediate step for protein synthesis. The genes encoded in the DNA are read and transcribed by the RNA polymerases (RNAP) into RNA, part of which, called messenger RNA, is subsequently translated into proteins by the ribosomes.
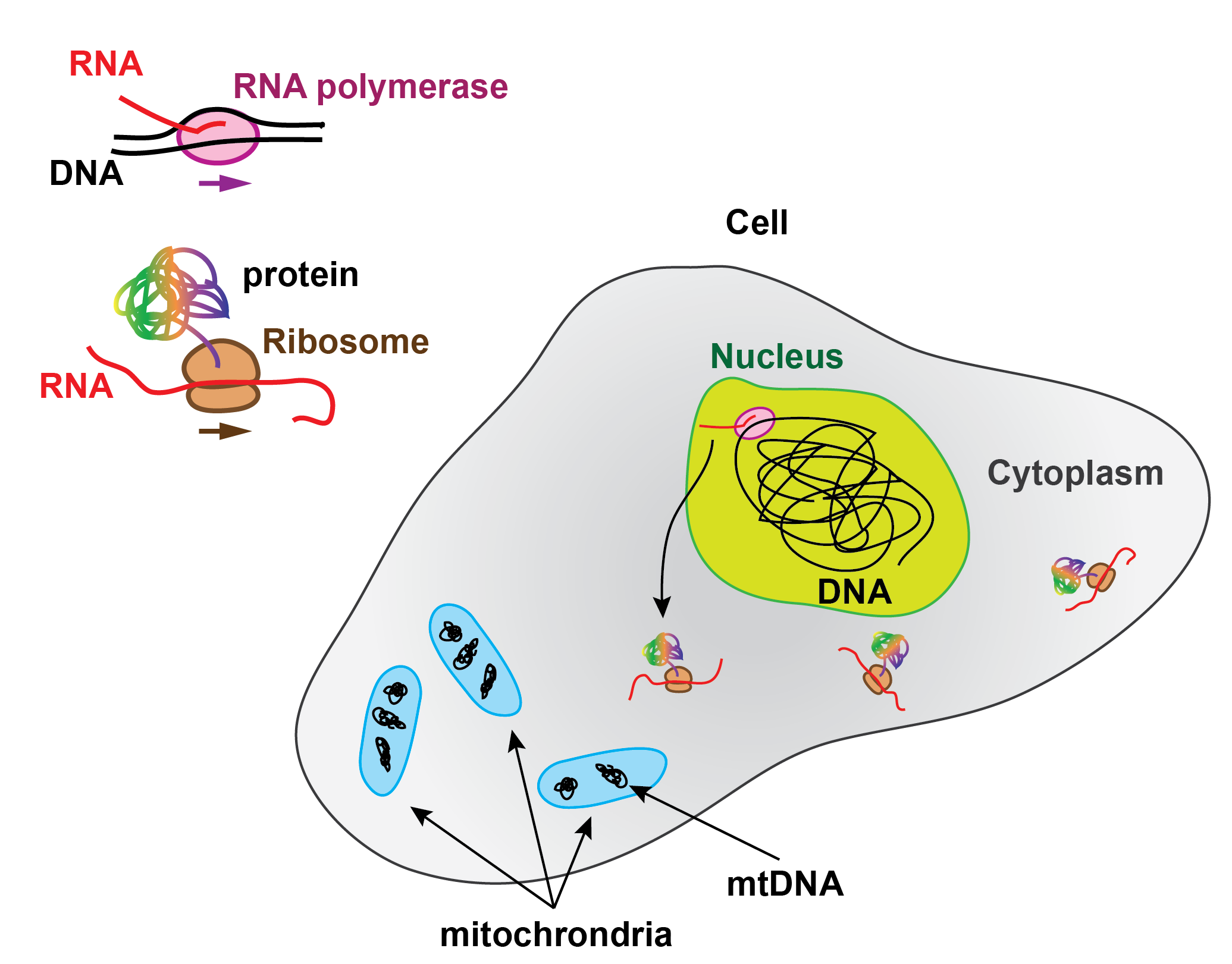
Schematic view of an eukaryotic cell and of the Central Dogma in molecular biology.
Being a key process in gene expression, transcription is therefore highly regulated in cells and is divided in three steps, i.e. initiation, elongation and termination.
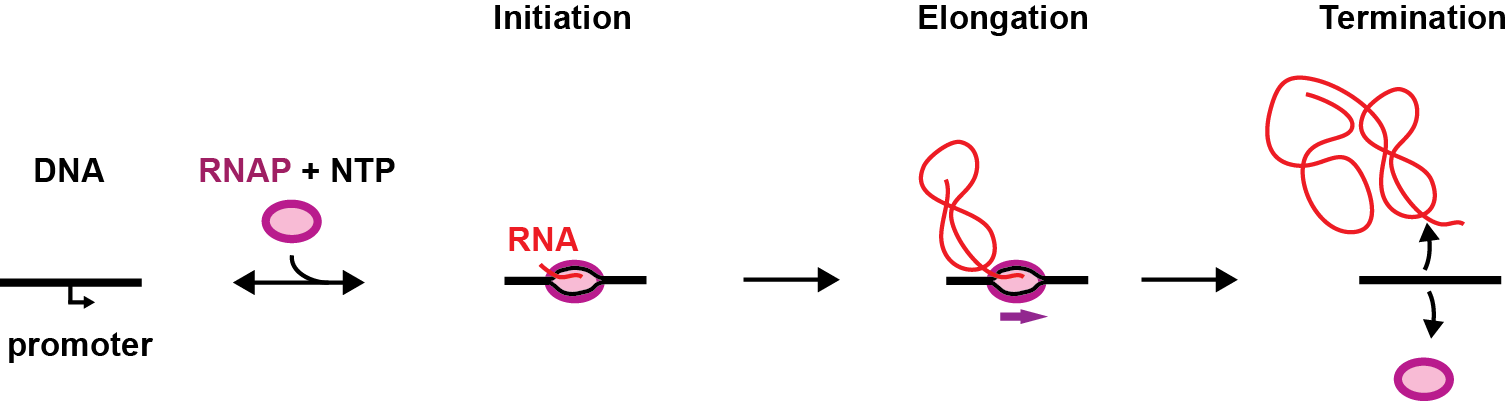
During initiation, the pre-initiation complex (PIC), formed by the RNAP and its associated factors, recognises the promoter, i.e. a DNA sequence that precedes the gene, and binds to it specifically, which forms the RNAP-promoter closed complex (RPc). The PIC subsequently opens the DNA promoter to make a transcription bubble, which forms the RNAP-promoter open complex (RPo) and places the RNAP active site at the transcription start site of the gene. In the presence of nucleotides (NTP), the RNAP initiates RNA synthesis (RPitc, itc: initially transcribing complex), which leads to either promoter escape towards elongation and dissociation from the PIC co-factors, or to abortive initiation, i.e. release of the nascent RNA, bringing the PIC back to the RPo stage. Interestingly, some promoters are rate-limiting for gene expression at the RPo stage, which is driven by promoter-PIC interactions, others at RPitc, driven by the initially transcribed sequence. Understanding the nature of these interactions and how they modulate promoter-based gene expression regulation is therefore of great importance.
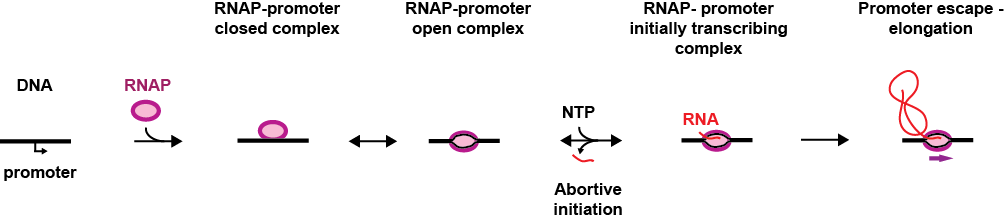
As every single steps during transcription initiation is reversible and stochastic, transcription initiation study using classical biochemistry ensemble experiments provides only the averaged behaviour of a heterogeneous sample. Therefore, ensemble techniques cannot access the kinetics of every intermediates, and provide an accurate description of transcription initiation. Single molecule techniques, such as single molecule FRET and magnetic tweezers, have been very useful to decipher the complex branched biochemical pathways of transcription initiation using their ability to observe the dynamics of individual RNA polymerases, and therefore discovered, e.g. pauses of various biochemical origins that regulates initial transcription in bacteria.
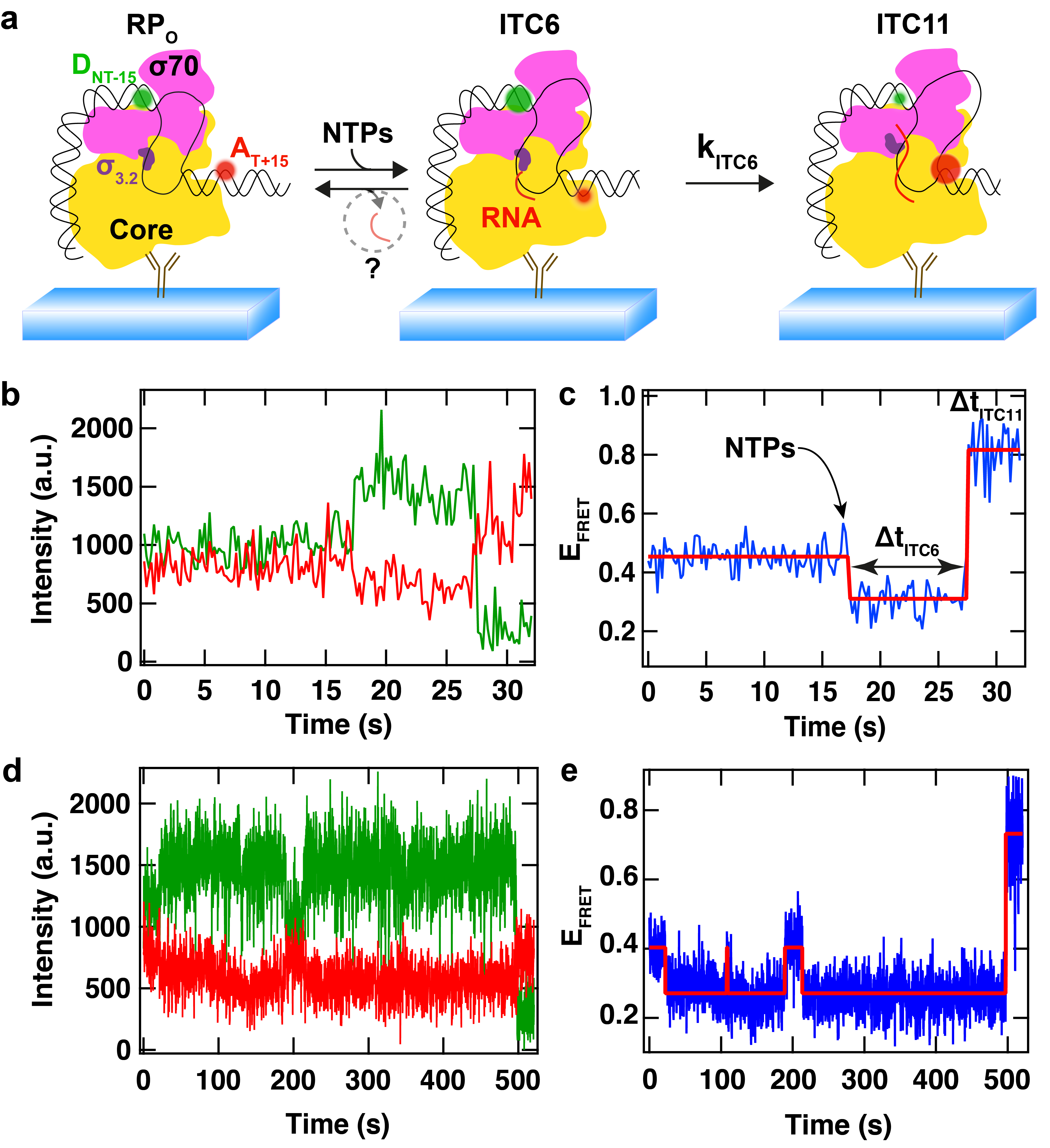
(a) Schematic of bacterial RNAP initial transcription study using single molecule FRET. The donor dye (D, green) and the acceptor dye (A, red) flank the transcription bubble. Upon NTP addition, the downstream DNA is scrunched in the RNAP, while the upstream DNA is static, which modifies the distance between the two dyes, leading to a change in FRET efficiency. (b) Fluorescence intensity of the donor (green) and of the acceptor (red) dyes. (c) FRET efficiency calculated from (b). The FRET efficiency (blue) first decreases upon addition of NTPs and the synthesis of the first 6 nucleotides of RNA (ITC6) by the RPitc, which pauses before extending the RNA to 11mer. The red step line is the Viterbi trace recovered from a Hidden Markov Model (HMM) to detect the changes in FRET efficiency. (d, e) Another FRET trace acquired in the same reaction conditions, though showing a completely different pausing behaviour. Adapted from Dulin et al., Nature Communications 2018.
In the Dulin lab, we investigate transcription initiation on various cellular systems, such as the human mitochondria transcription complex, using bespoke high throughput magnetic tweezers. We are currently developing a bespoke TIRF microscope for single molecule FRET experiments to complement our magnetic tweezers approach.
During elongation, it has been shown that the RNAP encounters various type of pauses that halt transcription. These pauses are important regulatory checkpoints for, e.g. transcription factor to bind the RNAP, to synchronise with co-transcriptional processes such as translation in bacteria. The high throughput capability of our magnetic tweezers assay offers the possibility to deeply probe transcription elongation and characterise rare – but relevant – biochemical states.
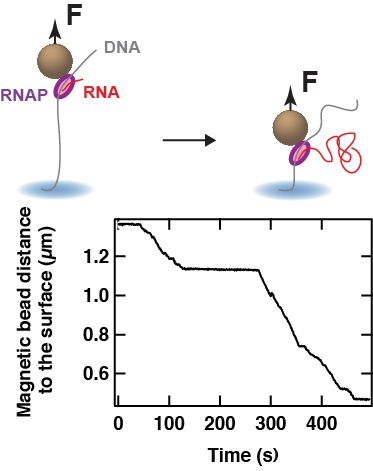
Elongating bacterial RNAP observation using a magnetic tweezers assay. The magnetic bead is bound to an RNAP that pulls the bead down against a constant force F while transcribing. The trace shows pauses of varying duration interrupting transcription.
At the end of the gene, the RNAP encounters a termination signal, which comes in the form of, e.g. DNA sequence that encodes a termination signal, such as an RNA hairpin, a protein complex that blocks and dissociates the RNAP. We are particularly interested in understanding protein roadblock mediated transcription termination, such as the one used to terminate human mitochondria transcription, where the mitochondrial termination factor 1 (MTERF1) promotes transcription termination in a directional manner.
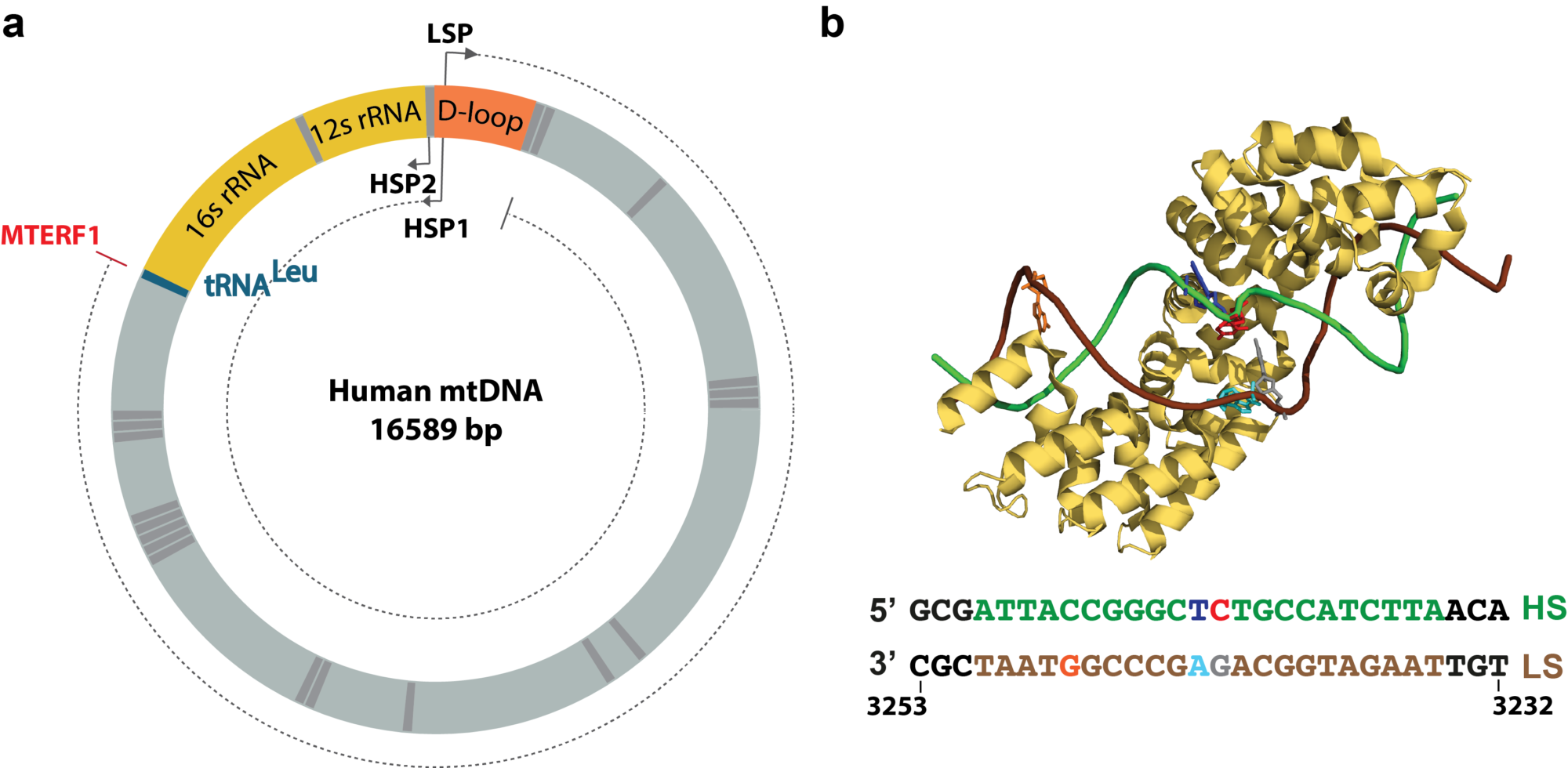
(a) Schematic of the human mitochondria genome, with the transcription promoters LSP, HSP1 and HSP2, and the termination binding site for MTERF1 positions indicated. MTERF1 blocks RNAP coming from LSP, but not from HSP1 and 2. (b) Structure of MTERF1 bound to the termination site, and termination site sequence with heavy and light strands (HS and LS, respectively) indicated.
(+)RNA virus replication
The Dulin lab has strong expertise in RNA virus replication study at the single molecule level, having pioneered the first studies of elongating viral RNA-dependent RNA polymerases (RdRp’s), the polymerase responsible for the synthesis of all viral genome. In particular, we focus our research on (+)RNA virus replication, i.e. virus of which the viral genome encodes directly the viral proteins. As any virus, (+)RNA viruses need the host cell translation machinery to synthesise the viral proteins from the 5-30 kilobases long viral genome.
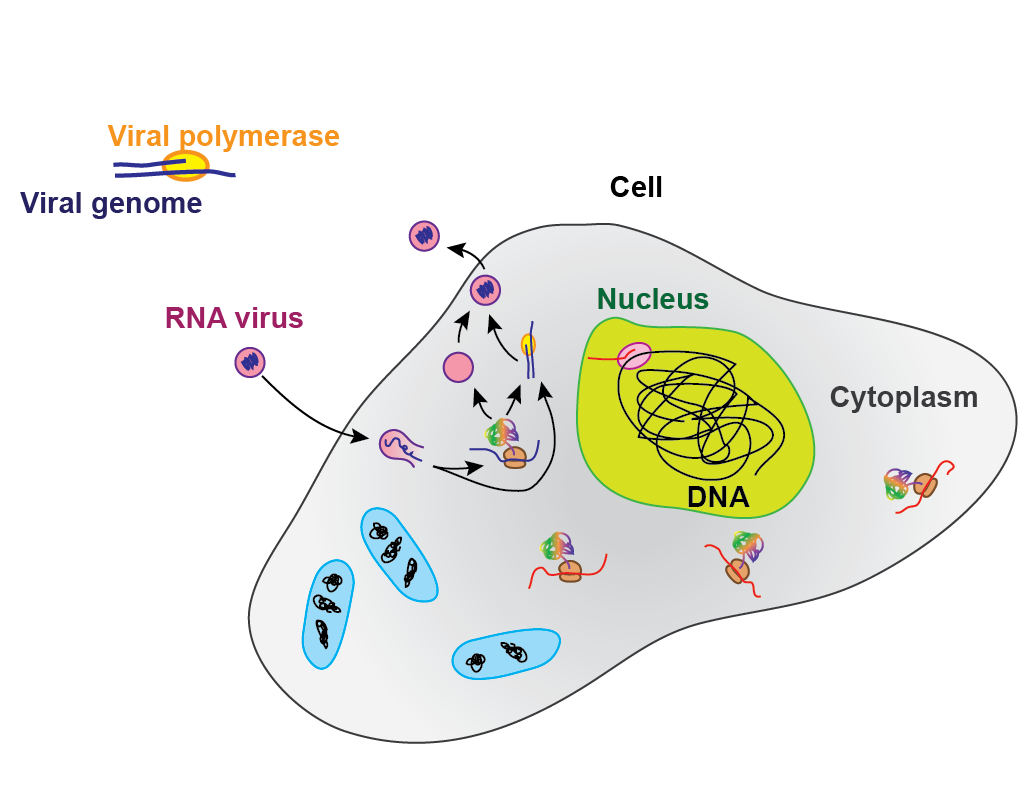
Schematic of the Central dogma in molecular biology, including (+)RNA virus genome processing.
Furthermore, the viral RdRp is also largely responsible for virus evolution and adaptation to the host antiviral defence mechanism, as it incorporates mutations in the viral genome and assist viral genome recombination, i.e. the mechanism by which part of the genome of two viruses are recombined to form a new viral genome. Given its importance, the viral RdRp is an important target for both drugs, e.g. antiviral nucleotide analogues, and vaccine development, e.g. by affecting the RdRp replication fidelity. Therefore, understanding viral replication kinetics and viral replication machinery interactions with the viral genome is of utmost importance. To characterise the mechanism of nucleotide misincorporation by the viral RdRp, we need a technique that allows the observation of rare (misincorporation probability ~0.001-0.0001/nucleotide incorporation) and asynchronous events. Furthermore, we need near single nucleotide spatial resolution and ~10-100ms temporal resolution, while using kilobases long template to collect enough miscincorporation events. High throughput magnetic tweezers are particularly suitable to solve this conundrum, as it offers the possibility to follow hundreds of polymerase simultaneously on kilobases long template, with ~0.3 nm tracking resolution at 1 s and constant applied force.
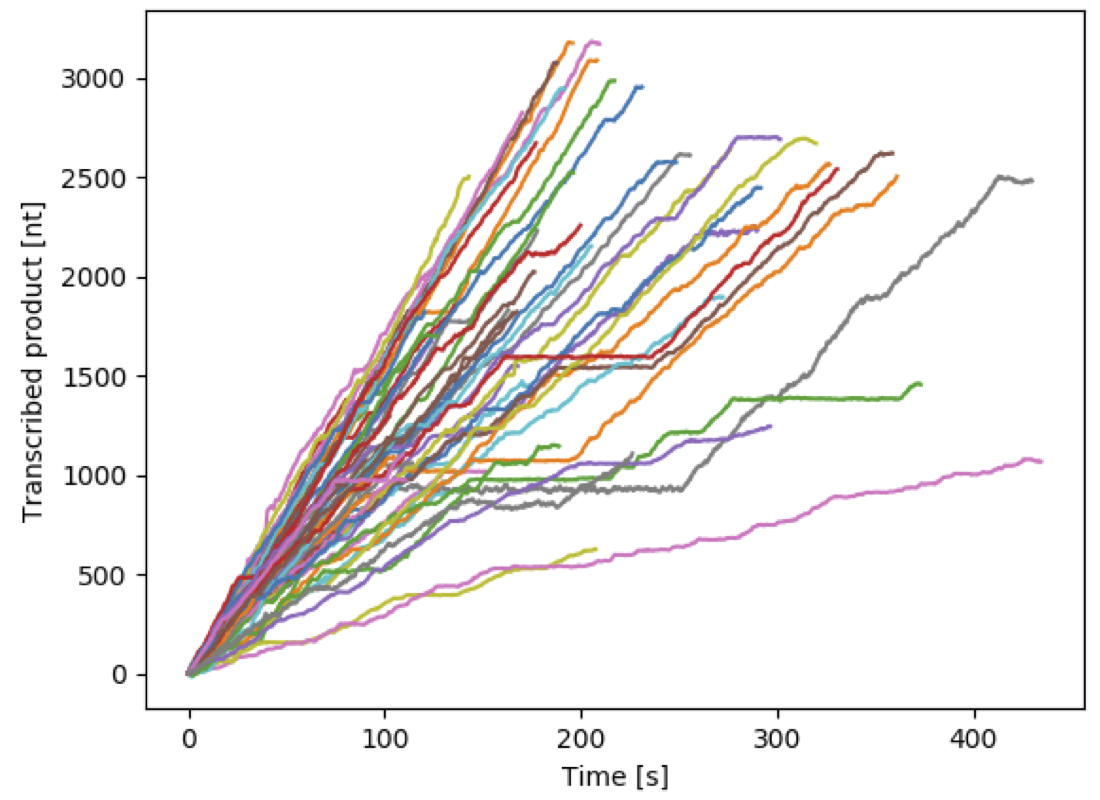
More than 100 viral RdRp traces acquired simultaneously in a single experiments using high-throughput magnetic tweezers. See experimental description here.
The study of the kinetics of viral RdRp’s using single molecule high throughput magnetic tweezers has been essential to discover a new mechanism for nucleotide misincorporation that generates a pause during viral genome replication (Dulin et al., Cell Reports 2015, 2017), but also to study how antiviral nucleotide analogues affect viral replication in the presence of all the natural nucleotides at saturating concentration, which is inaccessible for ensemble studies (as the antiviral nucleotide analogues are strongly selected against by the RdRp). This work has lead to the discovery of a new mode of action for antiviral nucleotides analogue, i.e. very long pauses linked to RdRp backtracking.
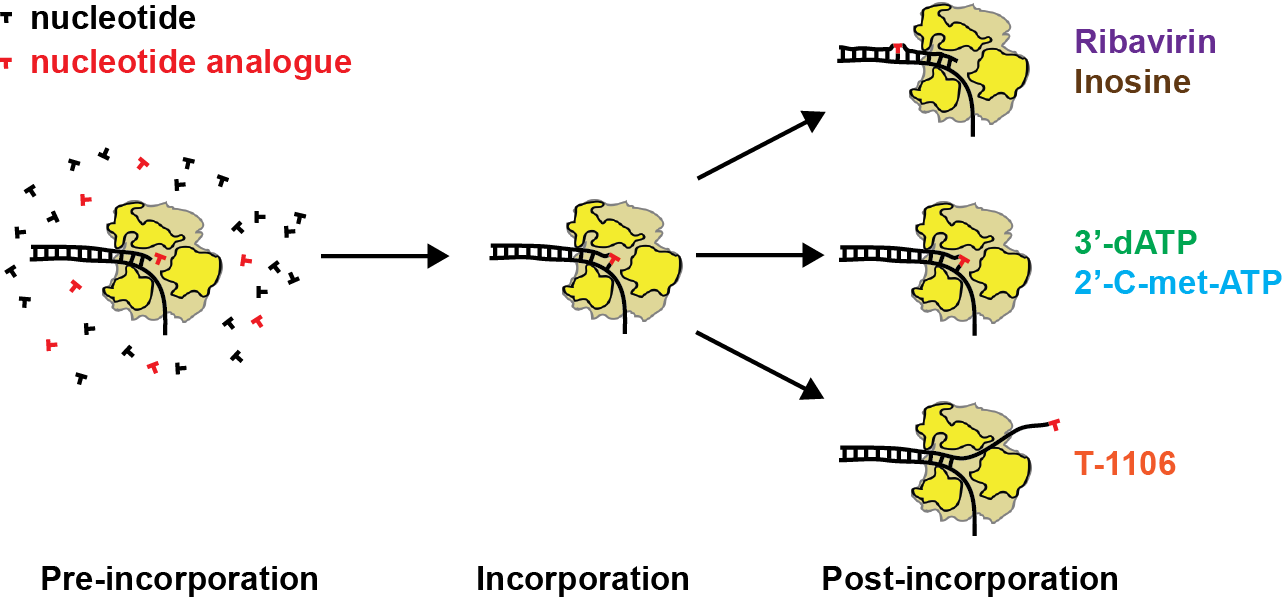
Nucleotide analogue effect on viral replication from single molecule studies: inosine and ribavirin induces a pause as misincorporation, 3′-dATP and 2′-C-met-ATP terminates replication and reduces RdRp processivity, and T1106 induces very long pauses related to RdRp backtracking.
The Dulin lab is now looking into how viral genome sequence affects viral replication kinetics, as well as how the viral genome interacts with the viral replication machinery. We particularly interested by the following virus genus, i.e. enterovirus (poliovirus, Rhinovirus-C), flavivirus (Dengue) and coronavirus (SARS).
